
Author, Programmer: Ole Lensmar (formerly Matzura), mailto:ole@lensmar.com
Co-Author: Anders Wennborg, mailto:anders.wennborg@cnt.ki.se
Last modified: June 18, 2012 (15 years after the previous update!)
This document outlines RNAdraw V1.01. A more thorough introduction and description of RNAdraw is available in the RNAdraw help files incorporated in the program.
All RNAdraw feedback will be gladly accepted at my above E-mail address!
Beta 2 Version of RNAdraw V1.1 available (Win32s support)!
2012-06-18: Download as ZIP file to be used under Windows XP/Vista/7/etc - rnadraw.zip
Index:
Latest Info (Check here on update and bug info)
System requirements and Installation

Welcome to RNAdraw! RNA structure prediction has in the past mainly been offered only on 'larger' computers due to the 16-bit memory restrictions implied by MS-DOS. With the Win32 API it is possible to port native 32-bit UNIX applications to Intel x86 compatible computers without implying any software based memory and calculation restrictions. Since Win32 is the base of Windows 95, Windows NT and Windows 3.1/3.11 with Win32s, ported programs should be able to run on an average Windows machine.
Most RNA structure prediction programs have previously had two restrictions:
1. They are available only on 'larger' computers, such as SUN, SGI and IBM systems.
2. They more or less lack a user-friendly interface.
RNAdraw issues both these problems and offers RNA optimal structure / basepair-probability matrix / heat curve calculation on Intel x86 compatible computers, providing a consistent user interface with many possibilities to view, print, import/export and edit calculation results.

To install and run RNAdraw on your system you will need the following:
- An Intel x86 (or compatible) computer (386, 486, P5, P6 )
- Windows 95, Windows NT or Window 3.1/3.11 with Win32s version 1.2 or later
- Newer versions of Windows (Vista, 7, etc) should work fine with the alternative ZIP file above
- at least 8Mb of RAM, the more the better
- 500 Kb free disk space
(Win32s is a subset of the Win32 API for Windows 3.1/3.11 available free from Microsoft or with RNAdraw.)
Memory requirements depend on sequence length and calculation type:
seq. size calc. structure calc. matrix --------------------------------------------------- 250 390 Kb 650 Kb 500 1530 Kb 2540 Kb 750 3410 Kb 5690 Kb 1000 6100 Kb 10100 Kb 1250 9440 Kb 15730 Kb 1500 13570 Kb 22620 Kb
As you can see, if you have 16 Mb of RAM, you can do up to 1000 bases easily (remember, the operating system wants some memory too..). On the other hand, if you lack sufficient memory for you calculations, you can do your calculations with Rnafold / GCG MFOLD / mfold on a UNIX machine and import the results into RNAdraw.
Currently there is no installation program for RNAdraw, you will have to install RNAdraw manually as described in the "install.txt" file included with RNAdraw.

Below you see a snapshot of RNAdraw V1.0 running under Windows 95:
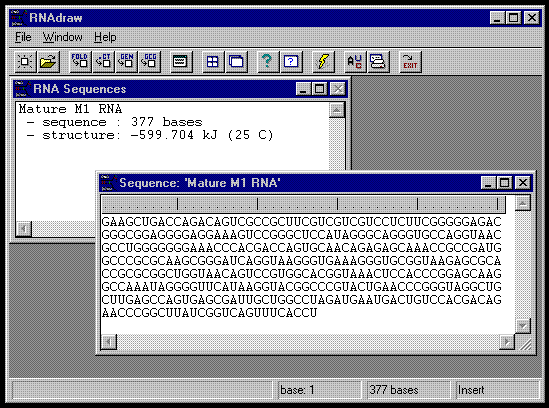
The sequence window ('RNA Sequences') is the 'control center' of RNAdraw. Here you see open files and their contents, such as structures and sequences. Double-clicking on an entry opens the corresponding edit window. As you can see above, the sequence entry for the first file has been opened. Here you can edit, cut and paste, import/export RNA sequences. Double-clicking on the sequence name in the list window opens up a notepad where you can maintain info about the file. You can for example import an E-mail into the notepad, cut out a sequence in the mail and paste it into the sequence editor. From there, you can do your structure /matrix /heat calculations and send them to the printer.
One very important concept in RNAdraw is right-button menus. All windows have an associated right-button menu, so does every entry in the sequence list. These menus all contain associated data manipulating functions (more on these below!).
RNAdraw supports 'drag-and-drop'; you can drag a .dra file from the File Manager/Explorer onto the main window and the file will be opened and added to the sequence list.
Extensive online help is also available.

The 'Calculate Structure(s)' menu option is available on the right-button menu of the sequence entry in the sequence list window. The minimum energy structure prediction algorithm in RNAdraw was ported from the Rnafold program included in the Vienna RNA package (1)
This dynamic programming algorithm is based on the work of M. Zuker and P. Stiegler (2) and uses energy parameters taken from Turner (3) , Freier (4) and Jaeger (5) . You can set multiple temperatures/temperature ranges for structure calculation, easily enter base-pairing constraints and set other calculation flags. The coordinates for the returned minimum energy structure are calculated with another ported program; Naview by R. Bruccoleri (6) .
Via a toolbar button or main menu option it is possible to modify/save/load all energy parameters used by the calculational algorithms in RNAdraw. In this way one can introduce new experimenatlly derived energies, rule out certain substructural motifs and monitor the effects of energy parameter changes on subsequently calculated structures.
Under Windows 95 and Windows NT, calculations can be done minimized in the background.

Below you see a snapshot of RNAdraw V1.0 running under Windows 95
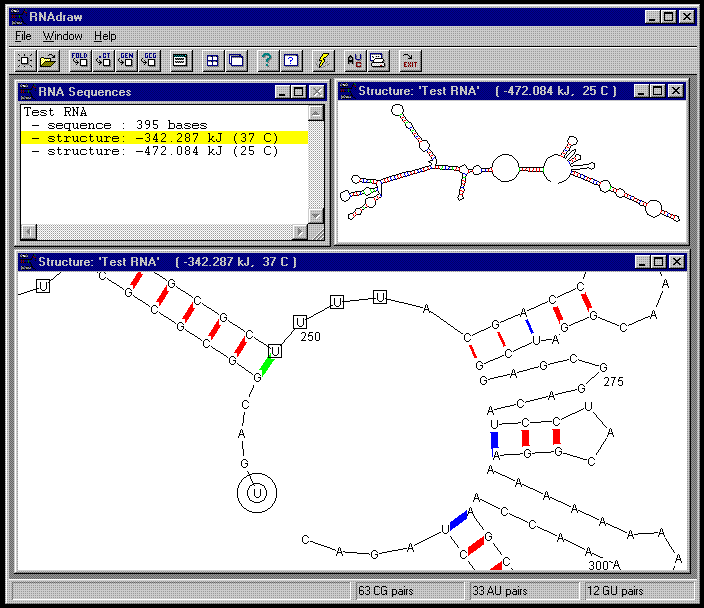
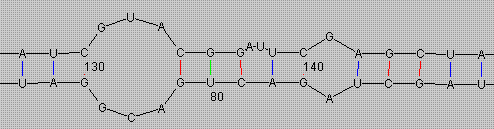
Above you see two structures in their edit windows. As you can see, you can adjust which information that will be shown, such as bases, labels, basepairs etc. You can easily zoom, pan and rotate the viewed structure by pressing the mouse buttons in different parts of the window, the mouse cursor changes appropriately to give you some notion of what is going to happen.
The associated right-button menu includes several manipulation functions. For example, 'Mark Bases' lets you search for and mark sequence tokens in numerous ways, as you can see in the bottom window where 'UUUU' was used as a token. 'Adjust Basepairs' allows the pairing / unpairing of individual basepairs to let you inspect structure / energy changes in different conformations.
It is possible to apply basepair probabilities to the structure view, resulting in thicker lines for higher probabilities, as you can see above.
All structure views can be printed on most Windows printers be exported as Windows bitmap (.bmp) or metafiles (.wmf/.emf).

The matrix calculation is either done in conjunction with structure calculation (see Structure Calculation ) or on its own. The partition function algorithm in RNAdraw was ported from the Rnafold program included in the Vienna RNA package (1) .
The matrix calculation is based on work by J.S McCaskill (7) , the energy parameters are taken from from Turner (3) , Freier (4) and Jaeger (5) . Just as with structure calculation, it is possible to set multiple temperatures/temperature ranges for matrix calculation, as well as other calculation flags.
Under Windows 95 and Windows NT, calculations can be done minimized in the background.

Below you see a snapshot of RNAdraw V1.0 running under Windows 95:
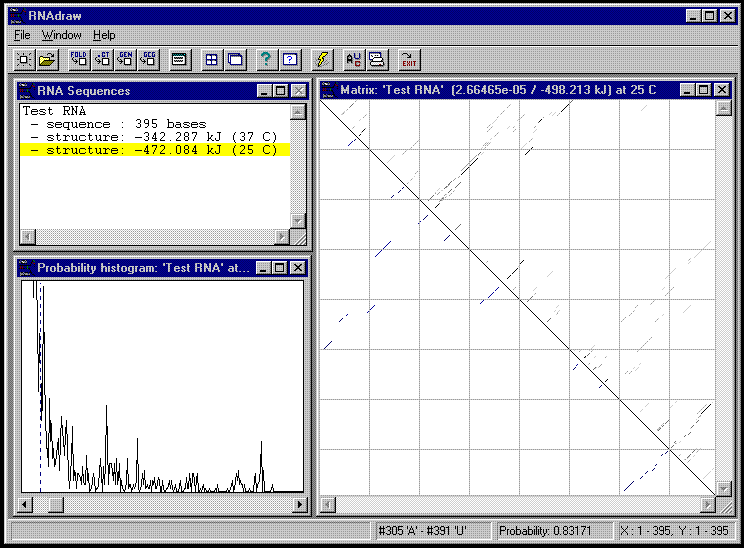
Above to the right you see the basepair-probability matrix of the 37C structure of the 'Random RNA'. This window can be opened from the 'Open Matrix' menu option, which is available on the structure view / structure entry right-button menu. The top right triangle shows the probability matrix, the bottom left triangle shows the minimum energy structure. As you can see, the probabilities are grayscale-coded (higher probability = darker gray). Is possible to zoom up and pan around with the scrollbars, show alignment lines etc.. Moving with the mouse over the matrix displays the base-pairing probabilities of the underlying base-pair, as you can see in the status bar.
You can manually pair/unpair basepairs in the structure by clicking with the left mouse button on a possible basepair in the lower left triangle. If you have the corresponding structure window open, you can directly monitor structure changes resluting from your basepair edits.
From the right-button menu of the matrix window you can open the matrix probability histogram window, which you see above on the bottom left. Here, you can see the probability frequency distribution of the matrix, print it if you want to, and set a cutoff frequency for which probabilities are shown in the matrix.
It is possible to 'extract' structures from the probability matrix, applying the current cutoff frequency. This allows you to view structures of certain 'probability levels', and compare them with thermodynamically optimal structures.
Both above windows can be exported and printed in the same manner as with structure views .

The 'Calculate Heat' menu option is available on the right-button menu of the sequence entry in the sequence list window. The specific heat calculation algorithm in RNAdraw was ported from the Rnaheat program included in the Vienna RNA package (1) .
The partition function algorithm is based on work by J.S McCaskill (7) , the energy parameters are taken from Turner (3) , Freier (4) and Jaeger (5) . You are able to set the temperature range and the temperature step value, as well as other calculation flags.
Under Windows 95 and Windows NT, calculations can be done minimized in the background (heat calculation can really take some time).

Below you see a snapshot of RNAdraw V1.0 running under Windows 95:
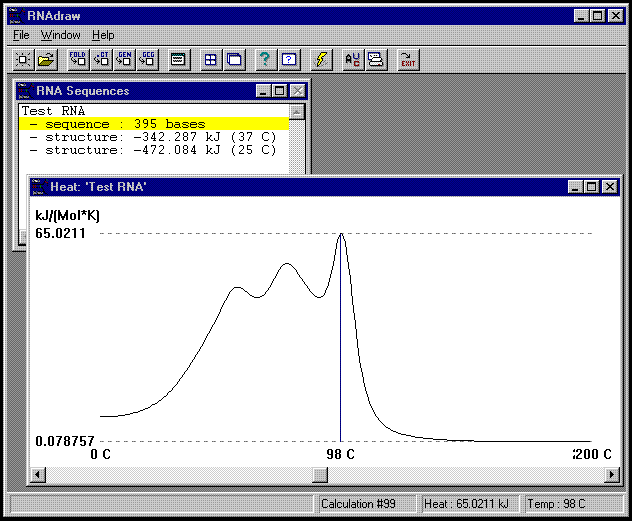
Above you see a Heat window. This window can be opened from the 'Show Heat' option, which is available on the sequence entry right-button menu. The scroll bar at the bottom of the window lets you move the above blue marker back and forth to see individual heat at specific temperatures in the status bar (as above).
You can print and export the window contents just as with the structure and matrix windows.

The following file formats can be imported into RNAdraw:
- Ms-DOS / UNIX text files into the notepad / sequence editor
- Any RNA text sequences can be pasted from the clipboard into the sequence editor.
- GENbank data files can be imported. The whole file will be inserted into the notepad, the sequence title will be parsed out and the sequence will be inserted into the sequence editor.
- Rnafold (1) output files can be used to create new sequence files since they both contain sequence and structure data. This allows you to do larger calculations on bigger machines and edit/view them on your PC.
- Rnafold (1) output files can also be merged with existing sequence files given that both files designate the same RNA sequence.
- Mulfold .ct files can be imported/merged in the same manner as Rnafold output files.
- GCG PlotFold -H files containing multiple structures can be imported into RNAdraw. Each structure will be in its own structure entry in the sequence list
The following data can be exported from RNAdraw:
- notepad info and sequences can be exported as Ms-DOS text files
- sequences can be exported as text files or copied to the clipboard
- all structure/matrix/heat windows can be printed and exported as:
1) Windows bitmap (.bmp) files
2) Windows standard / placeable / enhanced (not Win32s) metafiles

This has been, next to memory requirements, one of the main arguments against RNA structure prediction on the PC. Nowadays, 486 and especially Pentium machines deliver lots of calculation power, comparable to the capacity of IBM AS6000 systems. See the below table for some comparative results. The times shown are structure calculation/matrix calculation times in seconds.
( RNAdraw ) ( Rnafold )
size 486-66DX2 Pentium-90 AS 6000 SUN SPARC 10 (Dual proc.)
20 MB RAM 32 MB RAM 32 MB RAM 512 MB RAM
------------------------------------------------------------------------------
250 16/43 7/15 14/24 9/16
500 93/271 36/100 74/203 49/93
750 253/820 102/292 185/345 131/272
1000 510/1800 213/708 346/704 251/590
(results will vary between different machines)
As you can see, it doesn't take all to long time for smaller sequences on a 486-66DX2 computer, and surprisingly enough, a Pentium-90 outperforms even a SUN on structure calculation (remember though that this could well be due to better compiler output, not just processor horsepower).
The implemented version of NAVIEW (6) is extremly fast; the structure coordinates for a structure with 7250 bases are calculated within 4 seconds (!) on a 486-66DX2 computer.

I've got lots of plans for RNAdraw, but remember; if nobody seems to be using the program, I wont do much work on it. You don't have to register to encourage me, just send me a nice and constructive E-mail and I'll do what I can.
Planned calculation enhancements:
- multiple structure comparison, using both regional and local algorithms
- calculation of suboptimal structures using an improved prediction algorithm
Planned technical enhancements:
- Extended structure coordinate editing
- Modularity : The ability to plug in external algorithms into RNAdraw
- OLE 2.0 support
Have any ideas or comments? Mail me!

(1) I.L. Hofacker, W. Fontana, P.F. Stadler, L.S. Bonhoeffer, M. Tacker and P. Schuster, Chemical Monthly 125, 167-188 (1994)
(2) M. Zuker, P. Stiegler, Nucl. Acid. Res. 9: 133-148 (1981)
(3) D.H. Turner, N. Sugimoto and S.M. Freier, Ann. Rev. Biophys. Chem. 17: 167-192 (1988)
(4) S.M. Freier, R. Kiezerk, J.A. Jaeger, N. Sugimoto, M.H. Caruthers, T. Nelson, D.H. Turner, Proc. Nat. Acad. Sci 83, 9373-0377, 1986
(5) J.A. Jaeger, D.H. Turner, M. Zuker, Proc. Nat. Acad. Sci. 86, 7706-7710, 1989
(6) R. Bruccoleri, G. Heinrich, CABIOS 4, 167-173, (1988)
(7) J.S. McCaskill, Biopolymers 29, 1105-1119 (1990)

Check out the following: